Unraveling the Amplification-Free Quantitative Detection of Viral RNA in Nasopharyngeal Swab Samples Using a Compact Electrochemical Rapid Test Device
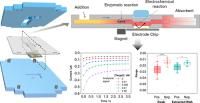
Providing viral load numbers of infection events aids in the identification of disease severity and in the effective overall patient management. Gold-standard polymerase chain reaction (PCR) techniques make this possible but cannot be applied at the point of need and in low-resource settings. Here, we report on the development of a compact analytical platform that can detect a conserved sequence of the RNA of severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) in 40 min in nasopharyngeal swab samples without the need for any previous purification or gene amplification steps. It combines electrochemical and paper fluidic approaches together with a sandwich hybridization assay performed on magnetic nanoparticles (MNPs) modified with a tailor-designed capture DNA hairpin. The device proves to quantitatively detect viral RNA in a retrospective study carried out with nasopharyngeal swab samples. A sensitivity of 100% and a specificity of 93% were estimated by the receiver operating characteristic (ROC) analysis. However, although molar concentration values of the target RNA sequence are provided, these estimates do not fully correlate with the viral load numbers estimated by RT-qPCR over the whole Ct sample range. Empirical studies have been carried out that have provided clear insights into this hurdle and simple solutions to overcome it, without depriving the device of the features required for potential use in a point-of-care (PoC) environment.
Anal. Chem. 2025, 97, 22, 11863–11873. DOI: 10.1021/acs.analchem.5c01605